Engineering Protein Assemblies
20.12.2019
Design of a new allosteric control system for protein assembly through the creation and rational modulation of a molecular switch.
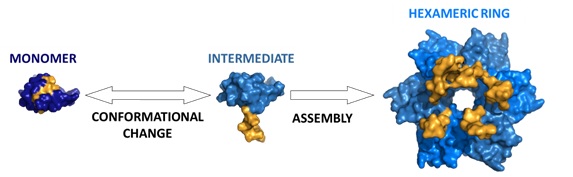
In nature there are numerous examples of biological macromolecules with allosteric control, i.e., where changes in one region of the molecule affect the behaviour and function in other regions of the molecule. However, the reproduction of this type of behaviour in synthetic molecules is very complex and so far the efforts invested in this area have only achieved a few successful results. In fact, in the specific case of the creation of new design assemblies by protein engineering, the results so far have been based on a puzzle system, where the surface of each of the rigid pieces to be assembled is modified to produce a stable interaction, without observing comformational changes. Now, researchers from IMDEA Nanociencia and the associated unit CNB-IMDEA, in collaboration with the Centro de Investigaciones Biológicas and the Universities of Granada and California have developed a model that allows to control in an allosteric way the formation and assembly of proteins through the inclusion of molecular switches formed thanks to changes in the three-dimensional structure of the protein.
The work, published in Nature Communications, demonstrates the potential of allosteric switches, using as a model the chymotrypsin inhibitor 2 of the barley seed. This protein, a protease inhibitor that protects the seed from being digested, is a small monomer protein that does not naturally oligomerate. The researchers, using protein engineering techniques, have introduced changes in the sequence of amino acids that produce the formation of oligomers (forming symmetrical rings of 6 and 12 subunits) through an extra step of control: a conformational change that acts as a switch that allows external control of how and when the protein oligomerizes. Luis Alberto Campos, first author of the work, explains that "this model system will serve as a basis for obtaining functional proteins with allosteric control of oligomerization, with possible applications in protein engineering, nanotechnology and synthetic biology." The use of these systems for modulating the transitions between monomer and oligomer provides greater control, dynamism and stability in the formation of macromolecular complexes and has applications in new research projects such as the creation of rings that interact with other molecules, such as metals or nucleic acids, or the control in the formation of membrane pores created by antimicrobial peptides through fusion proteins.
This work was supported by the European Research Council (grant ERC-2012-AdG-323059 to V.M.) and by the PRODESTECH network funded through the CONSOLIDER program from the Spanish Government (CSD2009-00088).
Keywords: Computational biophysics, Molecular engineering, Nanobiotechnology, Protein folding, Structural biology
Reference:
Engineering protein assembies with allosteric control via monomer fold-switching. Campos LA, Sharma R, Alvira S, Ruiz FM, Ibarra-Molero B, Sadqi M, Alfonso C, Rivas G, Sanchez-Ruiz JM, Romero-Garrido A, Valpuesta JM, Muñoz V. Nat Commun 2019 Dec 13;10(1):5703. doi: 10.1038/s41467-019-13686-1
Contact
IMDEA Nanociencia - Outreach Office
This email address is being protected from spambots. You need JavaScript enabled to view it.
+34 912 998712
Twitter: @imdea_nano
Facebook: @imdeananociencia
Source: CNB, IMDEA Nanociencia, Unidad Asociada de Nanobiotecnología IMDEA Nanociencia-CNB, Madrid, Spain